Current COBA Driving Biological Projects are listed below. Interested in becoming one of our Driving Biological Project partners? See our page on how to our collaborate with us or fill out our Project Inquiry form.
Principal Investigator |
Institution |
Project Description |
|
Robert Edwards |
University of California - San Francisco |
The Edwards lab studies the biochemistry and mechanisms of neurotransmission; this involves timing how long it takes for vesicles (such as those found at synapses) to release their cargo. We have worked with the lab to help identify and quantify exocytosis events in high-speed time-lapse movies. While previously identification of the events was manual, we have used Trackmate (link) alongside a Jupyter notebook to automatically detect the events and integrate these detections with the lab's own code for event classification. See our current workflow here. |
|
|
|
Jonathan Sexton |
University of Michigan |
The Sexton lab uses explanted pancreatic islets to study metabolic disorders and diabetes. These explants are difficult to image and analyze because they are 3D structures and because some cell types within the explants are rare but still need to be identified and quantified accurately. We worked with the lab to develop a pipeline in CellProfiler that utilizes a Cellpose deep learning model to segment nuclei and find cells in explants. See our current workflow here. To aid in classifying cells from these images into types, we've also added the ability to work with 3D datasets to CellProfiler Analyst. |
Charles Ndawula Jr |
National Livestock Resources Research Institute, Uganda |
Dr Ndawula studies tick-borne pathogens in livestock and develops methods to control their negative impact. Different species of ticks pose varying threats to their host, so determining the species of tick is essential. However, the identification of tick species can be nuanced, often subjective, and relies on visual guides as found here. This DBP aims to overcome the burden of understanding subtle details of ticks for new researchers by creating an intuitive deep learning-based tool for the classification of tick species for use while out in the field. | |
Jeff Hardin |
University of Wisconsin |
The Hardin Lab studies epidermal cell rearrangement in the C. elegans embryo using four-dimensional (4D) Nomarski microscopy. Previously, the lab relied on manual tracing of cells in these movies, making analysis and tracking slow. We’ve worked with the lab to create a FIJI script to find an in-focus plane on 4D datasets, and a Cellpose2 model to segment the epidermal cells. We're now working to create an automatic scoring of intercalation defects using TrackMate and improving the model to detect all cells in the embryo. | |
|
Amy Barczak |
Massachusetts General Hospital |
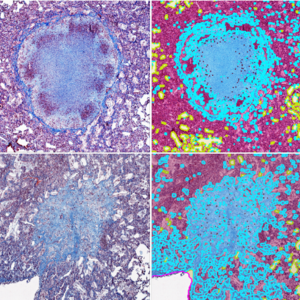
The Barczak lab studies fibrotic changes induced by tuberculosis using colorimetric staining on mouse lung tissue sections. For this project, we trained a robust pixel classifier that can detect collagen at an accuracy level comparable to human annotators despite the staining variabilities across batches. We developed a fully automated pipeline that identifies collagen, tissue, and air spaces within the whole slide images of lung tissue sections and filters the thinner strands of collagen to selectively provide a measure of “pathologic” collagen. |
Phil Newmark |
Morgridge Institute for Research |
The Newmark Lab uses planarians to study regeneration and germ development. We have worked with the lab to create workflows to align serial sections for 3D histopathology, challenging due to the planarians' small size. We have also created a number of workflows for 3D fluorescence imaging of testes, ovaries, and yolk glands. Initial fluorescence workflows made in CellProfiler have been improved to take advantage of Cellpose models. | |
Melissa Skala |
University of Wisconsin |
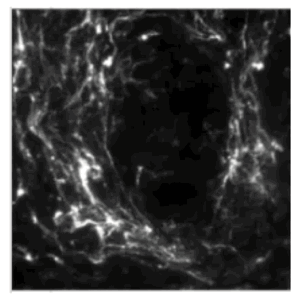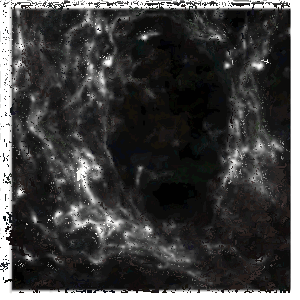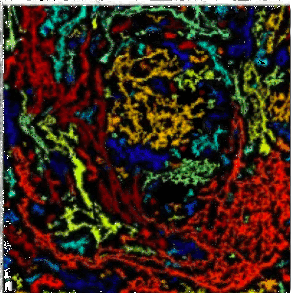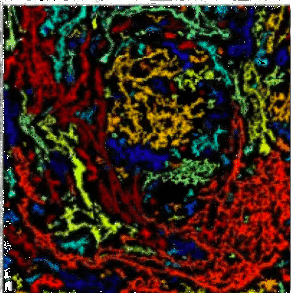
The Skala lab uses photonics-based technologies such as FLIM to develop personalized treatment plans for cancer patients (including breast, pancreatic, colorectal, neuroendocrine, oral, and other cancers). Because of the image modality, the number of cell types and the different cell culture conditions (2D and 3D), automated segmentation has been historically challenging. We have used CellPose 2.0 and CellProfiler to train deep learning models to segment the nuclei/cell and cytoplasm in a collection of cell types and then perform measurements on the segmented objects. | |
|
Thomas Gaborski |
Rochester Institute of Technology |
The Gaborski lab studies cell-substrate interactions, specifically how cells plated on different substrates secrete ECM molecules like collagen to generate fibers visualized with fluorescence microscopy. These images are challenging to analyze because the fibers form irregular, nonconvex shapes and are very heterogeneous in brightness. We've worked with the lab to generate a workflow using machine learning in ilastik and CellProfiler to segment fibers in these images. This single model works well across a variety of different fiber types and imaging setups. We've written up a blog post detailing this project here. |
Bill Bement |
University of Wisconsin |
The Bement Lab studies cytoskeletal organization and signaling. To help them detect particular patterns in microscopy images, we have developed several CellProfiler plugins, including plugins for scrambling pixels to destroy pre-existing structures and calculating the optimal variance sizes and variance metrics within an image. | |
Amy Shaub Maddox |
University of North Carolina - Chapel Hill |
The Maddox lab examines the intricate cellular structures involved in cell division using a variety of model systems. One such system is C. elegans, which the Maddox lab uses to study oogenesis with a desire to understand various characteristics of oocytes, such as their position and shape in 3D. These 3D images are difficult to segment since there is no nuclear stain and each cell is only defined by an intense staining at the cell borders. Previous methods used by the lab relied entirely on manual analysis which is time-consuming and low-throughput. This DBP aims to solve this by having an automated workflow for the segmentation and analysis of cell shape, size, and position. Materials for CellProfiler and deep-learning approaches are available here. | |
Chris Doe |
University of Oregon |
The Doe Lab studies central nervous system development in Drosophila, and is interested in tracing neurites to calculate metrics like branching and volume. Currently, this is only possible to the degree of accuracy needed with manual tracing. We are working to automate the segmentation of 3D images of neurites using deep learning techniques. | |

 |
Mary Harrington |
Smith College |
The Harrington Lab uses bioluminescent markers to study circadian rhythms in suprachiasmatic nuclei (SCN). These intravital images are challenging to segment because of the difficulty of aligning cells/regions between time points since the marker/signal can remain off for hours. We are working to align the images through the entire data set before the segmentation. |
